Computational Tools
CNL Research
Independent Component Analysis (ICA)
My lab developed the infomax ICA algorithm (Bell and Sejnowski 1995), which blindly separates mixtures of signals into component sources and has been used in hundreds of applications. We also pioneered the use of ICA to eliminate artifacts and analyze brains signals in EEG, MEG, fMRI and optical recording data (Delorme, Sejnowski et al. 2007; Siegel, Duann et al. 2007; Makeig, Gramann et al. 2009). Despite the thousands of papers that have used ICA to analyze brain data, it has so far not been possible to validate the brain sources directly. In collaboration with William Frost at Rosiland Franklin University, we have now confirmed the ability of ICA to isolate single neurons from optical recordings with simultaneous electrical recording in Tritonia and Aplysia, as shown in Fig. 4 (Hill, Moore-Kochlacs et al. 2010). Recent algorithmic improvements in ICA from my lab include generalization to Independent Vector Analysis (Hao, Lee et al. 2010), complex ICA for oscillating sources (Anemuller, Duann et al. 2006), and applications include sleep EEG analysis (Low, Shank et al. 2008), color vision (Wachtler, Doi et al. 2007) and speech recognition (Hao, Attias et al. 2009; Hao, Lee et al. 2010).
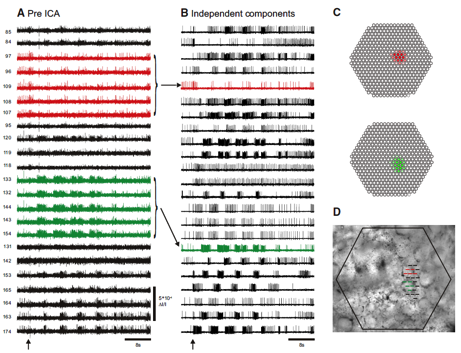
Fig. 1 Infomax Independent Component Analysis (ICA) (Bell and Sejnowski 1995) extracts the spiking activity of individual neurons from mixed and redundant photodiode array optical recording data. A: pre-ICA; 23 traces of a 464 trace photodiode array recording of a 4 cycle swim motor program in the dorsal pedal ganglion of Tritonia, elicited by a stimulus to pedal nerve (arrow). Typical of such recordings, neurons detected by many diodes are recorded redundantly (e.g., red and green traces), whereas many diodes detect signals from more than one neuron, resulting in signal mixtures. B: post-ICA; 23 of the 88 neuronal independent components returned by ICA containing recognizable neural activity. The independent components shown in red and green illustrate how redundancy in the optical recordings is eliminated by ICA. C: Contribution of each diode to the red and green components shown in B. D: the array locations of all 23 traces shown in A on the pedal ganglion (Hill, Moore-Kochlacs et al. 2010).
MCell – Monte Carlo Cell Model
MCell is a Monte Carlo computer program that simulates the biochemical reactions inside cells and the communication between neurons at synapses. Reactions often take place in spatial nanodomains, especially on the short time scales of synaptic signaling, and there are often only a handful of receptors and signal molecules. MCell tracks every signaling molecule in the synapse and incorporates realistic 3-D geometries obtained from EM reconstructions. In 2007, we released MCell3, a new version that incorporates reactions between diffusing molecules. We have incorporated new data structures for handling macromolecular complexes with an arbitrarily large number of internal states (Kerr, Bartol et al. 2008) (http://mcell.cnl.salk.edu/). MCell provides systems biology with a platform for simulating cellular interactions in 3D and is has been used in published papers by over 29 labs worldwide. MCell is not restricted to modeling neurons and we have used it to study cell division in bacteria (Kerr, Levine et al. 2006). We also collaborated recently with Roger Tsien to model the calcium concentration around the pores of ion channels (Tour, Adams et al. 2007) and with Mark Ellisman on modeling electrodiffusion at the node of Ranvier (Lopreore, Bartol et al. 2008).
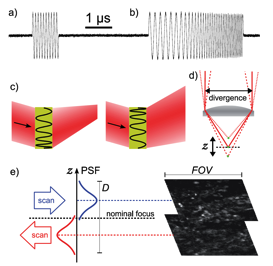
Fig. 2 Acousto-optic deflection (AOD) enables 3D scanning with no moving parts. a) Precise digital control of radio-frequency signals at microsecond timescales, b) Accurate and exactly repeatable sweeping between preset frequencies. c) A frequency sweep makes an AOD act as a lens, so the output beam can be made to converge or diverge depending on the direction of the sweep. d) Axial displacement of the focus from the nominal focal plane of the objective. e) Large volumes can be scanned rapidly through bidirectional line sweeps. The images show two focal planes, displaced by 40 μm in z, of EYFP-labeled neurons in mouse olfactory bulb taken simultaneously without moving the objective (Vucinic and Sejnowski 2007).
Optical Recording
We constructed a simple and compact imaging system designed specifically for the recording of fast neuronal activity in a 3D volume (Vucinic and Sejnowski 2007). The system uses an Yb:KYW femtosecond laser we designed for use with acousto-optic deflection. An integrated two-axis acousto-optic deflector, driven by digitally synthesized signals, can target locations in three dimensions. Data acquisition and the control of scanning were performed by a LeCroy digital oscilloscope. The total cost of construction was one order of magnitude lower than that of a typical Ti:sapphire system. The entire imaging apparatus, including the laser, is compact, low cost and simple to operate. The design offers easy integration with electrophysiology. The software and firmware is available for download at http://neurospy.org/ under an open source license.
References:
- Anemuller, J., J. R. Duann, et al. (2006). "Spatio-temporal dynamics in fMRI recordings revealed with complex independent component analysis." Neurocomputing 69(13-15): 1502-1512.
- Bell, A. J. and T. J. Sejnowski (1995). "An information-maximization approach to blind separation and blind deconvolution." Neural computation 7(6): 1129-1159.
- Delorme, A., T. Sejnowski, et al. (2007). "Enhanced detection of artifacts in EEG data using higher-order statistics and independent component analysis." NeuroImage 34(4): 1443-1449.
- Hao, J., H. Attias, et al. (2009). "Speech Enhancement, Gain, and Noise Spectrum Adaptation Using Approximate Bayesian Estimation." IEEE transactions on audio, speech, and language processing 17(1): 24-37.
- Hao, J., I. Lee, et al. (2010). "Independent vector analysis for source separation using a mixture of gaussians prior." Neural computation 22(6): 1646-1673.
- Hao, J., T. W. Lee, et al. (2010). "Speech Enhancement Using Gaussian Scale Mixture Models." IEEE transactions on audio, speech, and language processing 18(6): 1127-1136.
- Hill, E. S., C. Moore-Kochlacs, et al. (2010). "Validation of independent component analysis for rapid spike sorting of optical recording data." Journal of neurophysiology 104(6): 3721-3731.
- Kerr, R. A., T. M. Bartol, et al. (2008). "Fast Monte Carlo Simulation Methods for Biological Reaction-Diffusion Systems in Solution and on Surfaces." SIAM journal on scientific computing : a publication of the Society for Industrial and Applied Mathematics 30(6): 3126.
- Kerr, R. A., H. Levine, et al. (2006). "Division accuracy in a stochastic model of Min oscillations in Escherichia coli." Proceedings of the National Academy of Sciences of the United States of America 103(2): 347-352.
- Lopreore, C. L., T. M. Bartol, et al. (2008). "Computational modeling of three-dimensional electrodiffusion in biological systems: application to the node of Ranvier." Biophysical journal 95(6): 2624-2635.
- Low, P. S., S. S. Shank, et al. (2008). "Mammalian-like features of sleep structure in zebra finches." Proceedings of the National Academy of Sciences of the United States of America 105(26): 9081-9086.
- Makeig, S., K. Gramann, et al. (2009). "Linking brain, mind and behavior." International journal of psychophysiology : official journal of the International Organization of Psychophysiology 73(2): 95-100.
- Siegel, R. M., J. R. Duann, et al. (2007). "Spatiotemporal dynamics of the functional architecture for gain fields in inferior parietal lobule of behaving monkey." Cerebral cortex 17(2): 378-390.
- Tour, O., S. R. Adams, et al. (2007). "Calcium Green FlAsH as a genetically targeted small-molecule calcium indicator." Nature chemical biology 3(7): 423-431.
- Vucinic, D. and T. J. Sejnowski (2007). "A compact multiphoton 3D imaging system for recording fast neuronal activity." PloS one 2(1): e699.
- Wachtler, T., E. Doi, et al. (2007). "Cone selectivity derived from the responses of the retinal cone mosaic to natural scenes." Journal of vision 7(8): 6.
